Understanding tissue samples at the genetic level
The characterization of tissue samples using next-generation sequencing technology is a powerful tool that provides researchers with a new level of control. Without this genetic characterization, researchers have historically selected samples based on histological, and pathology data alone. Screening for the mutations of interest after the cohort has been received is both expensive and inefficient as only a portion of the samples will contain the desired mutation. It can take weeks to months of up-front work to conduct a genomic screening project such as this and it will result in a large volume of unwanted samples that could have been sourced for other more relevant projects.
SpecimenSeq NGS characterized FFPE samples
Discovery Life Sciences has genetically profiled thousands of diseased tissue samples using its SpecimenSeq platform. This additional layer of genomic information enables researchers to purchase sets of samples with known mutations, ensuring that 100% of the samples in a study population possess the required mutations, and eliminating the need to genetically characterize samples before starting a study, saving time, reducing costs, and accelerating research.
Using SpecimenSeq, Discovery Life Sciences has characterized over one and a half thousand different FFPE oncology tissue samples and identified over 800,000 variants of interest across a broad set of oncology indications. A data set that continues to grow as month after month more samples are genotyped and added to the Discovery database. The data collected includes specific information on oncogene mutations such as single nucleotide variant (SNV), fusions, deletions, substitutions, insertions, individual oncogene copy numbers, as well as tumor mutation burden (TMB), and microsatellite instability (MSI). Providing an unprecedented level of insight into the genetic makeup of these samples.
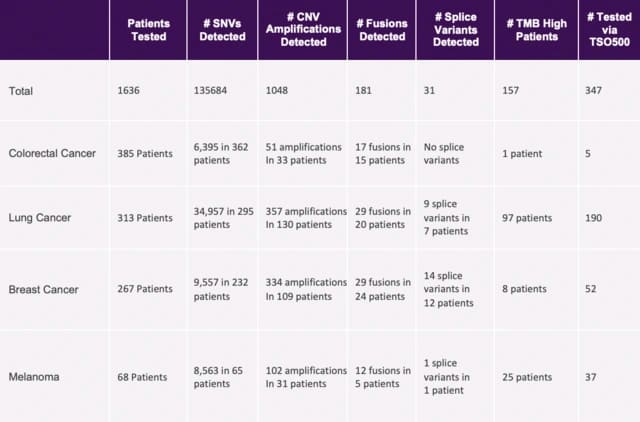
Table 1 shows a high-level snapshot of the available data, including the total number of data points across the total SpecimenSeq population of samples, as well as details for several common types of cancer. In total Discovery has accumulated data on over 90 different cancer indications using SpecimenSeq. Data that can be used to help researchers refine their cohorts and select populations of samples for narrowly defined cancer indications that include specific mutations.
Smart sample selection
Using SpecimenSeq, clients can work with Discovery’s in-house pathologists to identify a pool of samples ranging from single samples up to many thousands, all with specific pathology, histology, and genetic characteristics.
SpecimenSeq gives Discovery an unparalleled insight into the patterns and trends present across FFPE sample populations. One such insight is the prevalence of a mutation within a particular cancer indication. For a variety of reasons, that we will discuss in more depth in the next blog in this series, the actual prevalence of a mutation within the pool of accessible FFPE samples is typically different from the published prevalence in the general population. This can make a big difference e.g., when determining the number of lung cancer samples that will need to be screened to identify a cohort of 100 samples with a specific mutation. This information allows Discovery to respond much more rapidly to larger and more complex sample requests. In a situation where a customer needs a cohort of 150 diseased samples with a specific mutation and 50 healthy controls, Discovery may be able to supply 80 of the disease samples and all the controls immediately and will be able to accurately estimate, based on the known prevalence rates within similar FFPE populations, how long it will take to screen and identify the remaining 70 diseased samples. This level of accuracy is not available to a traditional biobank that lacks an in-house tissue and genomic biomarker service laboratories that can generate extensive genetic data on its samples.
Discovery scientists have first-hand experience selecting cohorts with specific biomarkers and understanding the true prevalence rates within the available samples. Depending on the size and complexity of the cohort required, Discovery may have all the samples in-house today with no requirement for a sequencing project, and if not, Discovery can accurately determine how long and how many samples will need to be screened to complete the cohort. Discovery works with customers using their consultative, client-centric, Science at Your Service approach that expertly helps identify and procure anywhere from a few samples to thousands from specific indication and within particular cancer indications.
Samples available with Specimen Seq data include FFPE, FFPE with matched plasma, DTCs, and BMMCs. You can learn more about genetically pre-characterized samples and SpecimenSeq on the Discovery Life Sciences website.

